Contributing to Sherpa development
Contributions to Sherpa - whether it be bug reports, documentation updates, or new code - are highly encouraged. Please report any problems or feature requests on github.
At present we do not have any explicit documentation on how to contribute to Sherpa, but it is similar to other open-source packages such as AstroPy.
The developer documentation is also currently lacking.
To do code development, Sherpa needs to be installed from source so that tests can run locally and the documentation can be build locally to test out any additions to code or docs. Building from source describes several ways to build Sherpa from source, but one particularly comfortable way is described in detail in the next section.
Pull requests
We welcome pull requests on github.
For each pull request, a set of continuous integration tests is run automatically, including a build of the documentation on readthedocs.
Skip the continuous integration
Sometimes a PR is still in development and known to fail the tests or
simply does not touch any code, because it only modifies docstrings
and the documentation. In that case, [skip ci] can be added to the
commit message to prevent running the github actions tests to save
time, energy, and limited resources.
Run tests locally
Before you issue a pull request, we ask to run the test suite locally.
Assuming everything is set up to install Sherpa from source, it can be
installed in development mode with pip:
pip install -e .
“Development mode” means that the tests will pick up changes in the
Python source files without running pip again (which can take some
time). Only if you change the C++ code, you will have to explicitly run
the installation again to see the changes in the tests. After the installation,
pytest can run all the tests. In the sherpa root directory call:
pytest
pytest supports a number of options which are
detailed in the pytest documentation. A
particularly useful option is to run only the tests in a specific file.
For example, if you changed the code and the tests in the sherpa.astro.ui
module, one might expect tests for this module to be the most likely to fail:
pytest sherpa/astro/ui/tests/test_astro_ui.py
Once everything looks good, you can do a final run of the entire test suite. A
second option useful for develoment is --pdb which drops into the
interactive Python debugger
when a test fails so that you can move up and down the stack and inspect the
value of individual variables.
The test suite can be sped up by running tests in parallel. After installing
the pytest-xdist module
(pip install pytest-xdist), tests can be run in parallel on several cores:
pytest -n auto
will autoselect the number of cores, an explicit number can also be given
(pytest -n 4). Note that if you have DS9 and XPA
installed then it is possible that the DS9 tests may fail when running
tests in parallel (since multiple tests can end up over-writing the
DS9 data before it can be checked).
Test coverage can be included as part of the tests by installing the
coverage
(pip install coverage) and
pytest-cov packages
(pip install pytest-cov). Adding the --cov=sherpa option to the test
run allows us to generate a coverage report after that:
pytest --cov=sherpa
coverage html -d report
The report is in report/index.html, which links to individual
files and shows exactly which lines were excuted while running the tests.
Run doctests locally
If doctestplus is installed
(and it probably is because it’s part of
sphinx-astropy,
which is required to build the documentation locally),
examples in the documentation are run automatically.
This serves two purposes:
it ensure that the examples we give are actually correct and match the code,
and it acts as additional tests of the Sherpa code base.
The doctest_norecursedirs setting in the pytests.ini file is used to exclude files which can not be
tested. This is generally because the examples were written before doctestplus support was added, and so
they need to be re-worked, or there is too much extra set-up required that would make the examples
hard-to follow. The file should be removed from this list when it has been updated to allow testing with doctestplus.
During development, you can run doctestplus on individual files like so (the option to use depends on whether it is a Python or reStructuredText file):
pytest --doctest-plus sherpa/astro/data.py
pytest --doctest-plus sherpa/data.py
pytest --doctest-rst docs/quick.rst
pytest --doctest-rst docs/evaluation/combine.rst
If you fix examples to pass these tests, remove them from the exclusion list in
pytest.ini! The goal is to eventually pass on all files.
Some doctests (in the documentation or in the docstrings of individual
functions) load data files. Those datafiles can be found in the
sherpa-test-data directory
as explained in the description of the development build.
There is a conftest.py file in the sherpa/docs directory and in the sherpa/sherpa
directory that sets up a
pytest fixture to define a variable called data_dir which points to this directory.
That way, we do not need to clutter the example with long directory names, but the
sherpa-test-data directory has to be present as a submodule to successfully pass all
doctests.
How do I …
Install from source in conda
Conda can be used to install all the dependencies for Sherpa, including XSPEC.
conda create -n sherpaciao -c https://cxc.cfa.harvard.edu/conda/ciao -c conda-forge ds9 astropy ciao
conda install -n sherpaciao --only-deps -c https://cxc.cfa.harvard.edu/conda/ciao -c conda-forge sherpa
conda activate sherpaciao
The first line installs the full CIAO release and astropy, required for building and running tests locally.
If you want to also build the documentation then add (after you have activated the environment):
conda install pandoc
pip install sphinx graphviz sphinx-astropy sphinx_rtd_theme nbsphinx ipykernel
Note
Sherpa can be configured to use crates (from CIAO) or astropy for
it’s I/O backend by changing the contents of the file
.sherpa-standalone.rc in your home directory. This file can be
found, once CIAO is installed, by using the get_config
routine:
% python -c 'import sherpa; print(sherpa.get_config())'
/home/happysherpauser/.sherpa-standalone.rc
If Sherpa was installed as part of CIAO then the file will be
called .sherpa.rc.
The io_pkg line in this file can be changed to select
crates rather than pyfits which would mean that astropy
does not need to be installed (although it would be needed to build
the documentation).
As described in Building from source, the file setup.cfg in
the root directory of the sherpa source needs to be modified to
configure the build. This is particularly easy in this setup, where
all external dependencies are installed in conda and the enviroment
variable ASCDS_INSTALL (or CONDA_PREFIX, which has the same
value) can be used. For most cases, the scripts/use_ciao_config
script can be used:
% ./scripts/use_ciao_config
Found XSPEC version: 12.12.0
Updating setup.cfg
% git diff setup.cfg
...
Otherwise the file can be edited manually. First find out what XSPEC version is present with:
% conda list xspec-modelsonly --json | grep version
"version": "12.12.0"
then change the setup.cfg to change the following lines, noting
that the ${ASCDS_INSTALL} environment variable must be
replaced by its actual value, and the xspec_version line
should be updated to match the output above:
bdist_wheel = sherpa_config xspec_config bdist_wheel
install_dir=${ASCDS_INSTALL}
configure=None
disable-group=True
disable-stk=True
fftw=local
fftw-include_dirs=${ASCDS_INSTALL}/include
fftw-lib-dirs=${ASCDS_INSTALL}/lib
fftw-libraries=fftw3
region=local
region-include_dirs=${ASCDS_INSTALL}/include
region-lib-dirs=${ASCDS_INSTALL}/lib
region-libraries=region ascdm
region-use-cxc-parser=True
wcs=local
wcs-include-dirs=${ASCDS_INSTALL}/include
wcs-lib-dirs=${ASCDS_INSTALL}/lib
wcs-libraries=wcs
with-xspec=True
xspec_version = 12.12.0
xspec_lib_dirs = ${ASCDS_INSTALL}/lib
xspec_include_dirs = ${ASCDS_INSTALL}/include
Note
The XSPEC version may include the patch level, such as 12.12.0e,
and this can be included in the configuration file.
To avoid accidentially commiting the modified setup.cfg into git,
the file can be marked as “assumed unchanged”.
git update-index --assume-unchanged setup.cfg
After these steps, Sherpa can be built from source:
pip install .
Warning
Just like in the case of a normal source install, when building Sherpa on recent versions of macOS within a conda environment, the following environment variable must be set:
export PYTHON_LDFLAGS=' '
That is, the variable is set to a space, not the empty string.
Warning
This is not guaranteed to build Sherpa in exactly the same manner as used by CIAO. Please create an issue if this causes problems.
Update the Zenodo citation information
The sherpa.citation() function returns citation information
taken from the Zenodo records for Sherpa.
It can query the Zenodo API, but it also contains a list of known
releases in the sherpa._get_citation_hardcoded routine. To add
to this list (for when there’s been a new release), run the
scripts/make_zenodo_release.py script with the version number
and add the screen output to the list in _get_citation_hardcoded.
For example, using release 4.12.2 would create (the author list has been simplified):
% ./scripts/make_zenodo_release.py 4.12.2
add(version='4.12.2', title='sherpa/sherpa: Sherpa 4.12.2',
date=todate(2020, 10, 27),
authors=['Doug Burke', 'Omar Laurino', ... 'Todd'],
idval='4141888')
Add a new notebook
The easiest way to add a new notebook to the documentation is to
add it to the desired location in the docs/ tree and add it to
the table of contents. If you want to place the notebook into the
top-level notebooks/ directory and also have it included in
the documentation then add an entry to the notebooks/nbmapping.dat
file, which is a tab-separated text file listing the name
of the notebook and the location in the docs/ directory structure
that it should be copied to. The docs/conf.py file will ensure
it is copied (if necessary) when building the documentation. The
location of the documentation version must be added to the
.gitignore file (see the section near the end) to make sure it
does not accidentally get added.
If the notebook is not placed in notebooks/ then the
nbsphinx_prolog setting in docs/conf.py will need updating.
This sets the text used to indicate the link to the notebook on the
Sherpa repository.
At present we require that the notebook be fully evaluated as we do not run the notebooks while building the documentation.
Add a new test option?
The sherpa/conftest.py file contains general-purpose testing
routines, fixtures, and configuration support for the test suite.
To add a new command-line option:
add to the
pytest_addoptionroutine, to add the option;add to
pytest_collection_modifyitemsif the option adds a new mark;and add support in
pytest_configure, such as registering a new mark.
Update the XSPEC bindings?
The sherpa.astro.xspec module currently supports
XSPEC versions 12.13.1, 12.13.0, 12.12.1, and 12.12.0. It may
build against newer versions, but if it does it will not provide
access to any new models in the release. The following sections of the
XSPEC manual
should be reviewed: Appendix F: Using the XSPEC Models Library in
Other Programs,
and Appendix C: Adding Models to XSPEC.
The spectral/manager/model.dat file provided by XSPEC - normally
in the parent directory of the HEADAS environment variable - defines
the interface for the models. The Sherpa module could be automatically
generated from this file but it would not be as informative as
manual generation (in particular the documentation), although this
could be changed (see the discussion at
issue #52).
Checking against a previous XSPEC version
If you have a version of Sherpa compiled with a previous XSPEC version then you can use three helper scripts:
scripts/check_xspec_update.pyThis will compare the supported XSPEC model classes to those from a
model.datfile, and report on the needed changes.scripts/update_xspec_functions.pyThis will report the text needed to go between the:
// Start model definitions ... // End model definitions
lines of the
sherpa/astro/xspec/src/_xspec.ccfile. This information is replicated in the output ofadd_xspec_model.pyso it depends on how many models need to be added or changed as to which to use.It is strongly suggested that the ordering from this routine is used, as it makes it easier to validate changes over time.
One issue is that this script can not identify which lines need be enclosed in a
#if def XSPEC_x_y_zblock, so care needs to be taken when updating the_xspec.ccfile with this output.scripts/add_xspec_model.pyThis will report the basic code needed to be added to both the compiled code (
sherpa/astro/xspec/src/_xspec.cc) and Python (sherpa/astro/xspec/__init__.py). Note that it does not deal with conditional compilation, the need to add a decorator to the Python class, or missing documentation for the class.
These routines are designed to simplify the process but are not guaranteed to handle all cases (as the model.dat file syntax is not strongly specified).
As an example of their use (the output will depend on the current Sherpa and XSPEC versions):
% ./scripts/check_xspec_update.py ~/local/heasoft-6.31/spectral/manager/model.dat | grep support
We do not support smaug (Add; xsmaug)
We do not support polconst (Mul; polconst)
We do not support pollin (Mul; pollin)
We do not support polpow (Mul; polpow)
We do not support pileup (Acn; pileup)
Note
There can be other output due to parameter-value changes
which are also important to review but this is just focussing
on the list of models that could be added to
sherpa.astro.xspec.
The screen output may differ slightly from that shown above, such as including the interface used by the model (e.g. C, C++, FORTRAN).
The list of function definitions, needed in _xspec.cc, can be
generated:
% ./scripts/update_xspec_functions.py ~/local/heasoft-6.31/spectral/manager/model.dat
XSPECMODELFCT_C_NORM(C_agauss, 3), // XSagauss
XSPECMODELFCT_NORM(agnsed, 16), // XSagnsed
XSPECMODELFCT_NORM(agnslim, 15), // XSagnslim
XSPECMODELFCT_C_NORM(C_apec, 4), // XSapec
...
XSPECMODELFCT_CON(C_zashift, 1), // XSzashift
XSPECMODELFCT_CON(C_zmshift, 1), // XSzmshift
XSPECMODELFCT_C_NORM(beckerwolff, 13), // XSbwcycl
Please note that this output needs to be reviewed as it can not identify which lines are conditional on the XSPEC version.
Although the wdem model is included in the XSPEC models, here is
how the add_xspec_model.py script can be used for those models
noted as not being supported:
% ./scripts/add_xspec_model.py ~/local/heasoft-6.31/spectral/manager/model.dat wdem
# C++ code for sherpa/astro/xspec/src/_xspec.cc
// Includes
#include <iostream>
#include <xsTypes.h>
#include <XSFunctions/Utilities/funcType.h>
#define XSPEC_12_12_0
#define XSPEC_12_12_1
#define XSPEC_12_13_0
#include "sherpa/astro/xspec_extension.hh"
// Defines
void cppModelWrapper(const double* energy, int nFlux, const double* params,
int spectrumNumber, double* flux, double* fluxError, const char* initStr,
int nPar, void (*cppFunc)(const RealArray&, const RealArray&,
int, RealArray&, RealArray&, const string&));
extern "C" {
XSCCall wDem;
void C_wDem(const double* energy, int nFlux, const double* params, int spectrumNumber, double* flux, double* fluxError, const char* initStr) {
const size_t nPar = 8;
cppModelWrapper(energy, nFlux, params, spectrumNumber, flux, fluxError, initStr, nPar, wDem);
}
}
// Wrapper
static PyMethodDef Wrappers[] = {
XSPECMODELFCT_C_NORM(C_wDem, 8),
{ NULL, NULL, 0, NULL }
};
// Module
static struct PyModuleDef wrapper_module = {
PyModuleDef_HEAD_INIT,
"_models",
NULL,
-1,
Wrappers,
};
PyMODINIT_FUNC PyInit__models(void) {
import_array();
return PyModule_Create(&wrapper_module);
}
# Python code for sherpa/astro/xspec/__init__.py
class XSwdem(XSAdditiveModel):
"""XSPEC AdditiveModel: wdem
Parameters
----------
Tmax
beta
inv_slope
nH
abundanc
Redshift
switch
norm
"""
_calc = _models.C_wDem
def __init__(self, name='wdem'):
self.Tmax = XSParameter(name, 'Tmax', 1.0, min=0.01, max=10.0, hard_min=0.01, hard_max=20.0, units='keV')
self.beta = XSParameter(name, 'beta', 0.1, min=0.01, max=1.0, hard_min=0.01, hard_max=1.0)
self.inv_slope = XSParameter(name, 'inv_slope', 0.25, min=-1.0, max=10.0, hard_min=-1.0, hard_max=10.0)
self.nH = XSParameter(name, 'nH', 1.0, min=1e-05, max=1e+19, hard_min=1e-06, hard_max=1e+20, frozen=True, units='cm^-3')
self.abundanc = XSParameter(name, 'abundanc', 1.0, min=0.0, max=10.0, hard_min=0.0, hard_max=10.0, frozen=True)
self.Redshift = XSParameter(name, 'Redshift', 0.0, min=-0.999, max=10.0, hard_min=-0.999, hard_max=10.0, frozen=True)
self.switch = XSParameter(name, 'switch', 2, alwaysfrozen=True)
# norm parameter is automatically added by XSAdditiveModel
pars = (self.Tmax, self.beta, self.inv_slope, self.nH, self.abundanc, self.Redshift, self.switch)
XSAdditiveModel.__init__(self, name, pars)
This code then can then be added to
sherpa/astro/xspec/src/_xspec.cc and
sherpa/astro/xspec/__init__.py and then refined so that the tests
pass.
Note
The output from add_xspec_model.py is designed for XSPEC user
models, and so contains output that either is not needed or is
already included in the _xspec.cc file.
Updating the code
The following steps are needed to update to a newer version, and assume that you have the new version of XSPEC, or its model library, available.
Add a new version define in
helpers/xspec_config.py.Current version: helpers/xspec_config.py.
When adding support for XSPEC 12.12.1, the top-level
SUPPORTED_VERSIONSlist was changed to include the triple(12, 12, 1):SUPPORTED_VERSIONS = [(12, 12, 0), (12, 12, 1)]
This list is used to select which functions to include when compiling the C++ interface code. For reference, the defines are named
XSPEC_<a>_<b>_<c>for each supported XSPEC release<a>.<b>.<c>(the XSPEC patch level is not included).Note
The Sherpa build system requires that the user indicate the version of XSPEC being used, via the
xspec_config.xspec_versionsetting in theirsetup.cfgfile (as attempts to identify this value automatically were not successful). This version is the value used in the checks inhelpers/xspec_config.py.Add the new version to
sherpa/astro/utils/xspec.pyThe
models_to_compiledroutine also contains aSUPPORTED_VERSIONSlist which should be kept in sync with the version inxspec_config.py.Attempt to build the XSPEC interface with:
pip install -e . --verbose
This requires that the
xspec_configsection of thesetup.cfgfile has been set up correctly for the new XSPEC release. The exact settings depend on how XSPEC was built (e.g. model only or as a full application), and are described in the building XSPEC documentation. The most-common changes are that the version numbers of theCCfits,wcslib, andhdsplibraries need updating, and these can be checked by looking in$HEADAS/lib.If the build succeeds, you can check that it has worked by directly importing the XSPEC module, such as with the following, which should print out the correct version:
python -c 'from sherpa.astro import xspec; print(xspec.get_xsversion())'
It may however fail, due to changes in the XSPEC interface (unfortunately, such changes are often not included in the release notes).
Identify changes in the XSPEC models.
Note
The
scripts/check_xspec_update.py,scripts/update_xspec_functions.py, andscripts/add_xspec_model.pyscripts can be used to automate some - but unfortunately not all - of this.A new XSPEC release can add models, change parameter settings in existing models, change how a model is called, or even delete a model (the last case is rare, and may require a discussion on how to proceed). The XSPEC release notes page provides an overview, but the
model.datfile - found inheadas-<version>/Xspec/src/manager/model.dat(build) or$HEADAS/../spectral/manager/model.dat(install) - provides the details. It greatly simplifies things if you have a copy of this file from the previous XSPEC version, since then a command like:diff heasoft-6.26.1/spectral/manager/model.dat heasoft-6.27/spectral/manager/model.dat
will tell you the differences (this example was for XSPEC 12.11.0, please adjust as appropriate). If you do not have the previous version then the release notes will tell you which models to look for in the
model.datfile.The
model.datis an ASCII file which is described in Appendix C: Adding Models to XSPEC of the XSPEC manual. The Sherpa interface to XSPEC only supports models labelled asadd,mul, andcon(additive, multiplicative, and convolution, respectively).Each model is represented by a set of consecutive lines in the file, and as of XSPEC 12.11.0, the file begins with:
% head -5 heasoft-6.27/Xspec/src/manager/model.dat agauss 2 0. 1.e20 C_agauss add 0 LineE A 10.0 0. 0. 1.e6 1.e6 0.01 Sigma A 1.0 0. 0. 1.e6 1.e6 0.01 agnsed 15 0.03 1.e20 agnsed add 0
The important parts of the model definition are the first line, which give the XSPEC model name (first parameter), number of parameters (second parameter), two numbers which we ignore, the name of the function that evaluates the model, the type (e.g.
add), and then 1 or more values which we ignore. Then there are lines which define the model parameters (the number match the second argument of the first line), and then one or more blank lines. In the output above we see that the XSPECagaussmodel has 2 parameters, is an additive model provided by theC_agaussfunction, and that the parameters areLineEandSigma. Theagnsedmodel is then defined (which uses theagnsedroutines), but its 15 parameters have been cut off from the output.The parameter lines will mostly look like this: parameter name, unit string (is often
" "), the default value, the hard and then soft minimum, then the soft ahd hard maximum, and then a value used by the XSPEC optimiser, but we only care about if it is negative (which indicates that the parameter should be frozen by default). The other common variant is the “flag” parameter - that is, a parameter that should never be thawed in a fit - which is indicated by starting the parameter name with a$symbol (although the documentation says these should only be followed by a single value, you’ll see a variety of formats in themodel.datfile). These parameters are marked by setting thealwaysfrozenargument of theParameterconstructor toTrue. Another option is the “scale” parameter, which is labelled with a*prefix, and these are treated as normal parameter values.Note
The examples below may refer to XSPEC versions we no-longer support.
sherpa/astro/xspec/src/_xspec.ccCurrent version: sherpa/astro/xspec/src/_xspec.cc.
New functions are added to the
XspecMethodsarray, using macros defined insherpa/include/sherpa/astro/xspec_extension.hh, and should be surrounded by a pre-processor check for the version symbol added tohelpers/xspec_config.py.As an example:
#ifdef XSPEC_12_12_0 XSPECMODELFCT_C_NORM(C_wDem, 8), // XSwdem #endif
adds support for the
C_wDemfunction, but only for XSPEC 12.12.0 and later. Note that the symbol name used here is not the XSPEC model name (the first argument of the model definition frommodel.dat), but the function name (the fifth argument of the model definition):% grep C_wDem $HEADAS/../spectral/manager/model.dat wdem 7 0. 1.e20 C_wDem add 0
Some models have changed the name of the function over time, so the pre-processor directive may need to be more complex, such as the following (although now we no-longer support XSPEC 12.10.0 this particular example has been removed from the code):
#ifdef XSPEC_12_10_0 XSPECMODELFCT_C_NORM(C_nsmaxg, 6), // XSnsmaxg #else XSPECMODELFCT_NORM(nsmaxg, 6), // XSnsmaxg #endif
The remaining pieces are the choice of macro (e.g.
XSPECMODELFCT_NORMorXSPECMODELFCT_C_NORM) and the value for the second argument. The macro depends on the model type and the name of the function (which defines the interface that XSPEC provides for the model, such as single- or double- precision, and Fortran- or C- style linking). Additive models use the suffix_NORMand convolution models use the suffix_CON. Model functions which begin withC_use the_Cvariant, while those which begin withc_currently require treating them as if they have no prefix.The numeric argument to the template defines the number of parameters supported by the model once in Sherpa, and should equal the value given in the
model.datfile for multiplicative and convolution style models, and one larger than this for additive models (i.e. those which use a macro that ends in_NORM).As an example, the following three models from
model.dat:apec 3 0. 1.e20 C_apec add 0 phabs 1 0.03 1.e20 xsphab mul 0 gsmooth 2 0. 1.e20 C_gsmooth con 0
are encoded as (ignoring any pre-processor directives):
XSPECMODELFCT_C_NORM(C_apec, 4), // XSapec XSPECMODELFCT(xsphab, 1), // XSphabs XSPECMODELFCT_CON(C_gsmooth, 2), // XSgsmooth
The
scripts/update_xspec_functions.pyscript will create a list of all the supported models for the suppliedmodel.datfile, and can be used to fill up the text between the:// Start model definitions ... // End model definitions
markers. However, the script can not determine the state of each symbol (e.g. whether it needs to be protected by a version check, as discussed earlier), or to add further notes, so care needs to be taken to update this information.
Those models that do not use the
_Cversion of the macro (or, for convolution-style models, have to useXSPECMODELFCT_CON_F77), also have to declare the function within theextern "C" {}block. For FORTRAN models, the declaration should look like (replacingfuncwith the function name, and note the trailing underscore):xsf77Call func_;
and for model functions called
c_func, the prefixless version should be declared as:xsccCall func;
If you are unsure, do not add a declaration and then try to build Sherpa: the compiler should fail with an indication of what symbol names are missing.
Note
Ideally we would have a sensible ordering for the declarations in this file, but at present it is ad-hoc.
sherpa/astro/xspec/__init__.pyCurrent version: sherpa/astro/xspec/__init__.py.
This is where the Python classes are added for additive and multiplicative models. The code additions are defined by the model and parameter specifications from the
model.datfile, and the existing classes should be used for inspiration. The model class should be calledXS<name>, where<name>is the XSPEC model name, and thenameargument to its constructor be set to the XSPEC model name.The two main issues are:
Documentation: there is no machine-readable version of the text, and so the documentation for the XSPEC model is used for inspiration.
The idea is to provide minimal documentation, such as the model name and parameter descriptions, and then to point users to the XSPEC model page for more information.
One wrinkle is that the XSPEC manual does not provide a stable URI for a model (as it can change with XSPEC version). However, it appears that you can use the following pattern:
https://heasarc.gsfc.nasa.gov/xanadu/xspec/manual/XSmodel<Name>.html
where
<Name>is the capitalised version of the model name (e.g.Agnsed).Models that are not in older versions of XSPEC should be marked with the
version_at_leastdecorator (giving it the minimum supported XSPEC version as a string), and the function (added to_xspec.cc) is specified as a string using the__function__attribute. Thesherpa.astro.xspec.utils.ModelMetametaclass performs a runtime check to ensure that the model can be used.For example (from when XSPEC 12.9.0 was still supported):
__function__ = "C_apec" if equal_or_greater_than("12.9.1") else "xsaped"
sherpa/astro/xspec/tests/test_xspec.pyCurrent version: sherpa/astro/xspec/tests/test_xspec.py.
The
XSPEC_MODELS_COUNTversion should be increased by the number of models classes added to__init__.py.Additive and multiplicative models will be run as part of the test suite - using a simple test which runs on a default grid and uses the default parameter values - whereas convolution models are not (since their pre-conditions are harder to set up automatically).
docs/model_classes/astro_xspec.rstCurrent version: docs/model_classes/astro_xspec.rst.
New models should be added to both the
Classesrubric - sorted by addtive and then multiplicative models, using an alphabetical sorting - and to the appropriateinheritance-diagramrule.
Documentation updates
The
docs/indices.rstfile should be updated to add the new version to the list of supported versions, under the XSPEC term, anddocs/developer/index.rstalso lists the supported versions (Update the XSPEC bindings?). The installation pagedocs/install.rstshould be updated to add an entry for thesetup.cfgchanges in XSPEC.The
sherpa/astro/xspec/__init__.pyfile also lists the supported XSPEC versions.
Never forget to update the year of the copyright notice?
Git offers pre-commit hooks
that can do file checks for you before a commit is executed. The script in
scripts/pre-commit will check if the copyright notice in any of the files in the
current commit must be updated and, if so, add the current year to the copyright notice
and abort the commit so that you can manually check before commiting again.
To use this opt-in functionality, simply copy the file to the appropriate location:
cp scripts/pre-commit .git/hooks
Notes
Notes on the design and changes to Sherpa.
N-dimensional data and models
Models and data objects are
designed to work with flattened arrays, so a 1D dataset has x and
y for the independent and dependent axes, and a 2D dataset will
have x0, x1, and y values, with each value stored as a 1D
ndarray. This makes it easy to deal with filters and sparse or
irregularly-placed grids.
>>> from sherpa.data import Data1D, Data1DInt, Data2D
As examples, we have a one-dimensional dataset with data values (dependent axis, y) of 2.3, 13.2, and -4.3 corresponding to the independent axis (x) values of 1, 2, and 5:
>>> d1 = Data1D("ex1", [1, 2, 5], [2.3, 13.2, -4.3])
An “integrated” one-dimensional dataset for the independent axis bins 23-44, 45-50, 50-53, and 55-57, with data values of 12, 14, 2, and 22 looks like this:
>>> d2 = Data1DInt("ex2", [23, 45, 50, 55], [44, 50, 53, 57], [12, 14, 2, 22])
An irregularly-gridded 2D dataset, with points at (-200, -200), (-200, 0), (0, 0), (200, -100), and (200, 150) can be created with:
>>> d3 = Data2D("ex3", [-200, -200, 0, 200, 200], [-200, 0, 0, -100, 150],
... [12, 15, 23, 45, -2])
A regularly-gridded 2D dataset can be created, but note that the arguments must be flattened:
>>> import numpy as np
>>> x1, x0 = np.mgrid[20:30:2, 5:20:2]
>>> shp = x0.shape
>>> y = np.sqrt((x0 - 10)**2 + (x1 - 31)**2)
>>> x0 = x0.flatten()
>>> x1 = x1.flatten()
>>> y = y.flatten()
>>> d4 = Data2D("ex4", x0, x1, y, shape=shp)
The dimensionality of models
Originally the Sherpa model class did not enforce any requirement on the models, so it was possible to combine 1D and 2D models, even though the results are unlikely to make sense. With the start of the regrid support, added in PR #469, the class hierarchy included 1D- and 2D- specific classes, but there was still no check on model expressions. This section describes the current way that models are checked:
the
sherpa.models.model.Modelclass defines asherpa.models.model.Model.ndimattribute, which is set toNoneby default.the
sherpa.models.model.RegriddableModel1Dandsherpa.models.model.RegriddableModel2Dclasses set this attribute to 1 or 2, respectively (most user-callable classes are derived from one of these two classes).the
sherpa.models.model.CompositeModelclass checks thendimattribute for the components it is given (thepartsargument) and checks that they all have the samendimvalue (ignoring those models whose dimensionality is set toNone). If there is a mis-match then asherpa.utils.err.ModelErris raised.as described below, the dimensions of data and model can be compared.
An alternative approach would have been to introduce 1D and 2D specific classes, from which all models derive, and then require the parent classes to match. This was not attempted as it would require significantly-larger changes to Sherpa (but this change could still be made in the future).
The data class
Prior to Sherpa 4.14.1, the Data object did not have
many explicit checks on the data it was sent, instead relying on
checks when the data was used. Now, validation checks are done
when fields are changed, rather than when the data
is used. This has been done primarily by marking field accessors as
property attributes, so that they can apply the validation checks when
the field is changed. The intention is not to catch all possible
problems, but to cover the obvious cases.
Data dimensionality
Data objects have a ndim field,
which is used to ensure that the model and data dimensions match when
using the eval_model and
eval_model_to_fit methods.
The size of a data object
The size field describes the size of a data
object, that is the number of individual elements. Once a data object
has its size set it can not be changed (this is new to Sherpa 4.14.1,
as in previous versions you could change fields to any size). This
field can also be accessed using len, with it returning 0 when no
data has been set.
Point versus Integrated
There is currently no easy way to identify whether a data object requires integrated (low and high edges) or point axes (the coordinate at which to evaluate the model).
Handling the independent axis
Checks have been added in Sherpa 4.14.1 to ensure that the correct
number of arrays are used when setting the independent axis: that is,
a Data1D object uses (x,), Data1DInt
uses (lo, hi), and Data2D uses (x0, x1). Note that
the arguement is expected to be a tuple, even in the
Data1D case, and that the individual components are
checked to ensure they have the same size.
The handling of the independent axis is mediated by a “Data Space”
object (DataSpaceND, DataSpace1D,
IntegratedDataSpace1D, DataSpace2D, and
IntegratedDataSpace2D) which is handled by the
_init_data_space and _check_data_space methods of the
Data class.
To ensure that any filter remains valid, the independent axis is
marked as read-only. The only way to change a value is to change the
whole independent axis, in which case the code recognizes that the
filter - whether just the mask attribute or also
any region filter for the DataIMG case - has to
be cleared.
Validation
Fields are converted to ndarray - if not None - and then checked
to see if they are 1D and have the correct size. Some fields may have
extra checks, such as the grouping and
quality columns for PHA data which
are converted to integer values.
One example of incomplete validation is that the
bin_lo and
bin_hi fields are not checked to ensure
that both are set, or that they are in descending order, that the
bin_hi value is always larger than the correspondnig bin_lo
value, or that there are no overlapping bins.
Error messages
Errors are generally raised as DataErr exceptions,
although there are cases when a ValueError or TypeError will be
raised. The aim is to provide some context in the message, such as:
>>> from sherpa.data import Data1D
>>> x = np.asarray([1, 2, 3])
>>> y = np.asarray([1, 2])
>>> data = Data1D('example', x, y)
Traceback (most recent call last):
...
sherpa.utils.err.DataErr: size mismatch between independent axis and y: 3 vs 2
and:
>>> data = Data1D('example', x, x + 10)
>>> data.apply_filter(y)
Traceback (most recent call last):
...
sherpa.utils.err.DataErr: size mismatch between data and array: 3 vs 2
For DataPHA objects, where some length checks
have to allow either the full size (all channels) or just the filtered
data, the error messages could explain that both are allowed, but this
was felt to be overly complicated, so the filtered size will be used.
PHA Filtering
Filtering of a DataPHA object has four
complications compared to Data1D objects:
the independent axis can be referred to in channel units (normally 1 to the maximum number of channels), energy units (e.g. 0.5 to 7 keV), or wavelength units (e.g. 20 to 22 Angstroms);
each channel has a width of 1, so channel filters - which are generally going to be integer values - map exactly, but each channel has a finite width in the derived units (that is, energy or wavelength) so multiple values will map to the same channel (e.g. a channel may map to the energy range of 0.4 to 0.5 keV, so any value >= 0.4 and < 0.5 will map to it);
the data can be dynamically grouped via the
groupingattribute, normally set by methods likegroup_counts()and controlled by thegroup()method, which means that the desired filter, when mapped to channel units, is likely to end up partially overlapping the first and last groups, which means thatnotice(a, b)andignore(None, a); ignore(b, None)are not guaranteed to select the same range;and there is the concept of the
qualityarray, which defines whether channels should either always be, or can temporarily be, ignored.
This means that a notice() or
ignore() call has to convert from
the units of the input - which is defined by the
units attribute, changeable with
set_analysis - to the “group
number” which then gets sent to the
_data_space attribute to track
the filter.
One result is that the mask attribute
will now depend on the grouping scheme. The
get_mask method can be used to
calculate a mask for all channels (e.g. the ungrouped data).
There are complications to this from the quality concept introduced by the OGIP grouping scheme, which I have not been able to fully trace through in the code.
Combining model expressions
Models can be combined in several ways (for models derived from the
sherpa.models.model.ArithmeticModel class):
a unary operator, taking advantage of the
__neg__and__abs__special methods of a class;a binary operator, using the
__add__,__sub__,__mul__,__div__,__floordiv__,__truediv__,__mod__and__pow__methods.
This allows models such as:
sherpa.models.basic.Polynom1D('continuum') + sherpa.models.basic.Gauss1D('line')
to be created, and relies on the sherpa.models.model.UnaryOpModel
and sherpa.models.model.BinaryOpModel classes.
The BinaryOpModel class has special-case handling
for values that are not a model expression (i.e. that do not derive
from the ArithmeticModel class),
such as:
32424.43 * sherpa.astro.xspec.XSpowerlaw('pl')
In this case the term 32424.43 is converted to an
ArithmeticConstantModel instance and then
combined with the remaining model instance (XSpowerlaw).
For those models that require the full set of elements, such as
multiplication by a RMF or a convolution kernel, this requires
creating a model that can “wrap” another model. The wrapping model
will evaluate the wrapped model on the requested grid, and then apply
any modifications. Examples include the
sherpa.instrument.PSFModel class, which creates
sherpa.instrument.ConvolutionModel instances, and the
sherpa.astro.xspec.XSConvolutionKernel class, which
creates sherpa.astro.xspec.XSConvolutionModel instances.
When combining models, BinaryOpModel
(actually, this check is handled by the super class
CompositeModel), this approach will ensure that the
dimensions of the two expressions match. There are some models, such
as TableModel and
ArithmeticConstantModel, which do not
have a ndim attribute (well, it
is set to None); when combining components these are ignored, hence
treated as having “any” dimension.
Plotting data using the UI layer
The plotting routines, such as
plot_data() and
plot_fit(),
follow the same scheme:
The plot object is retrieved by the appropriate
get_xxx_plotroutine, such asget_data_plot()andget_fit_plot().These
get_xxx_plotcalls retrieve the correct plot object - which is normally a sub-class ofPlotorHistogram- from the session object.Note
The naming of these objects in the
Sessionobject is rather hap-hazard and would benefit from a more-structured approach.If the
recalcargument is set then thepreparemethod of the plot object is called, along with the needed data, which depends on the plot type - e.g.sherpa.plot.DataPlot.prepareneeds data and statistic objects andsherpa.plot.ModelPlot.prepareneeds data and model objects (and a statistic class too but in this case it isn’t used).Calls to other access other plot objects may be required, such as the fit plot requiring both data and model objects. It is also the place that specialised logic, such as selecting a histogram-style plot for
Data1DIntdata rather than the default plot style, is made.These plot objects generally do not require a plotting backend, so they can be set and returned even without Matplotlib installed.
Once the plot object has been retrieved, is is sent to a plotting routine -
sherpa.ui.utils.Session._plot()- which calls theplotmethod of the object, passing through the plot options. It is at this point that the plot backend is used to create the visualization (these settings are passed as**kwargsdown to the plot backend routines).
The sherpa.astro.ui.utils.Session class adds a number
of plot types and classes, as well as adds support for the
DataPHA class to relevant
plot commands, such as plot_model()
and plot_fit(). This
support complicates the interpretation of the model and fit types,
as different plot types are used to represent the model when drawn
directly (plot_model) and indirectly (plot_fit): these plot
classes handle binning differently (that is, whether to apply the
grouping from the source PHA dataset or use the native grid of the
response).
There are two routines that return the preference settings:
get_data_plot_prefs and
get_model_plot_prefs.
The idea for these is that they return the preference dictionary that
the relevant classes use. However, with the move to per-dataset
plot types (in particular Data1DInt and
DataPHA). It is not entirely clear
how well this scheme works.
The contour routines follow the same scheme, although there is a
lot less specialization of these methods, which makes the
implementation easier. For these plot objects the
sherpa.ui.utils.Session._contour() method is used
instead (and rather than have overplot we have overcontour
as the argument).
The sherpa.ui.utils.Session.plot() and
sherpa.ui.utils.Session.contour() methods allow multiple
plots to be created by specifying the plot type as a list of
argumemts. For example:
>>> s.plot('data', 'model', 'data', 2, 'model', 2)
will create four plots, in a two-by-two grid, showing the
data and model values for the default dataset and the
dataset numbered 2. The implementation builds on top of the
individual routines, by mapping the command value to the
necessary get_xxx_plot or get_xxx_contour routine.
The image routines are conceptually the same, but the actual
implementation is different, in that it uses a centralized
routine to create the image objects rather than have the
logic encoded in the relavant get_xxx_image routines. It is
planned to update the image code to match the plot and contour
routines. The main difference is that the image display is handled
via XPA calls to an external DS9 application, rather than with
direct calls to the plotting library.
As an example, here I plot a “fit” for a Data1DInt
dataset:
>>> from sherpa.ui.utils import Session
>>> from sherpa.data import Data1DInt
>>> from sherpa.models.basic import Const1D
>>> s = Session()
>>> xlo = [2, 3, 5, 7, 8]
>>> xhi = [3, 5, 6, 8, 9]
>>> y = [10, 27, 14, 10, 14]
>>> s.load_arrays(1, xlo, xhi, y, Data1DInt)
>>> mdl = Const1D('mdl')
>>> mdl.c0 = 6
>>> s.set_source(mdl)
>>> s.plot_fit()
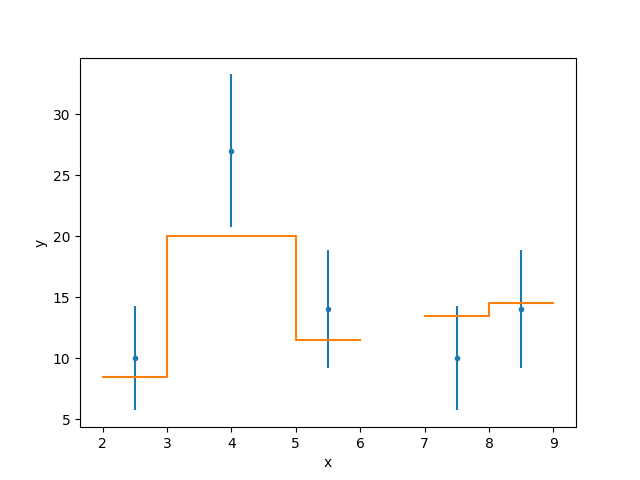
We can see how the Matplotlib-specific options are passed
to the backend, using a combination of direct access,
such as color='black', and via the preferences
(the marker settings):
>>> s.plot_data(color='black')
>>> p = s.get_model_plot_prefs()
>>> p['marker'] = '*'
>>> p['markerfacecolor'] = 'green'
>>> p['markersize'] = 12
>>> s.plot_model(linestyle=':', alpha=0.7, overplot=True)
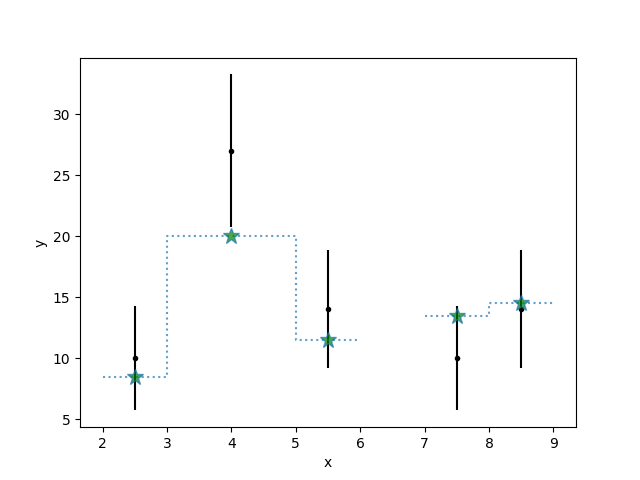
We can view the model plot object:
>>> plot = s.get_model_plot(recalc=False)
>>> print(type(plot))
<class 'sherpa.plot.ModelHistogramPlot'>
>>> print(plot)
xlo = [2,3,5,7,8]
xhi = [3,5,6,8,9]
y = [ 6.,12., 6., 6., 6.]
xlabel = x
ylabel = y
title = Model
histo_prefs = {'xerrorbars': False, 'yerrorbars': False, ..., 'linecolor': None}
Coordinate conversion for image data
The sherpa.data.Data2D class provides basic support for
fitting models to two-dimensional data; that is, data with two
independent axes (called “x0” and “x1” although they should be
accessed via the indep attribute). The
sherpa.astro.data.DataIMG class extends the 2D support to
include the concept of a coordinate system, allowing the independent
axis to be one of:
logicalimageworld
where the aim is that the logical system refers to a pixel number (no
coordinate system), image is a linear transform of the logical system,
and world identifies a projection from the image system onto the
celestial sphere. However, there is no requirement that this
categorization holds as it depends on whether the optional
sky and
eqpos attributes are set when
the DataIMG object is created.
Using a coordinate system directly
Note
It is expected that the DataIMG object
is used with a rectangular grid of data and a shape attribute
set up to describe the grid shape, as used in the next
section, but it is not required, as shown
here.
If the independent axes are known, and not calculated via a coordinate
transform, then they can just be set when creating the
DataIMG object, leaving the
coord attribute set to
logical.
>>> from sherpa.astro.data import DataIMG
>>> x0 = np.asarray([1000, 1200, 2000])
>>> x1 = np.asarray([-500, 500, -500])
>>> y = np.asarray([10, 200, 30])
>>> d = DataIMG("example", x0, x1, y)
>>> print(d)
name = example
x0 = Int64[3]
x1 = Int64[3]
y = Int64[3]
shape = None
staterror = None
syserror = None
sky = None
eqpos = None
coord = logical
This can then be used to evaluate a two-dimensional model,
such as Gauss2D:
>>> from sherpa.models.basic import Gauss2D
>>> mdl = Gauss2D()
>>> mdl.xpos = 1500
>>> mdl.ypos = -100
>>> mdl.fwhm = 1000
>>> mdl.ampl = 100
>>> print(mdl)
gauss2d
Param Type Value Min Max Units
----- ---- ----- --- --- -----
gauss2d.fwhm thawed 1000 1.17549e-38 3.40282e+38
gauss2d.xpos thawed 1500 -3.40282e+38 3.40282e+38
gauss2d.ypos thawed -100 -3.40282e+38 3.40282e+38
gauss2d.ellip frozen 0 0 0.999
gauss2d.theta frozen 0 -6.28319 6.28319 radians
gauss2d.ampl thawed 100 -3.40282e+38 3.40282e+38
>>> d.eval_model(mdl)
array([32.08564744, 28.71745887, 32.08564744])
Attempting to change the coordinate system with
set_coord will error out with a
DataErr instance reporting that the data
set does not specify a shape.
The shape attribute
The shape argument can be set when creating a
DataIMG object to indicate that the
data represents an “image”, that is a rectangular, contiguous, set of
pixels. It is defined as (nx1, nx0), and so matches the ndarray
shape attribute from NumPy. Operations that treat the dataset as a
2D grid often require that the shape attribute is set.
>>> x1, x0 = np.mgrid[1:4, 1:5]
>>> y2 = (x0 - 2.5)**2 + (x1 - 2)**2
>>> y = np.sqrt(y2)
>>> d = DataIMG('img', x0.flatten(), x1.flatten(),
... y.flatten(), shape=y.shape)
>>> print(d)
name = img
x0 = Int64[12]
x1 = Int64[12]
y = Float64[12]
shape = (3, 4)
staterror = None
syserror = None
sky = None
eqpos = None
coord = logical
>>> d.get_x0()
array([1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4])
>>> d.get_x1()
array([1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3])
>>> d.get_dep()
array([1.80277564, 1.11803399, 1.11803399, 1.80277564, 1.5 ,
0.5 , 0.5 , 1.5 , 1.80277564, 1.11803399,
1.11803399, 1.80277564])
>>> d.get_axes()
(array([1., 2., 3., 4.]), array([1., 2., 3.]))
>>> d.get_dims()
(4, 3)
Attempting to change the coordinate system with
set_coord will error out with a
DataErr instance reporting that the data
set does not contain the required coordinate system.
Setting a coordinate system
The sherpa.astro.io.wcs.WCS class is used to add a
coordinate system to an image. It has support for linear (translation
and scale) and “wcs” - currently only tangent-plane projections
are supported - conversions.
>>> from sherpa.astro.io.wcs import WCS
>>> sky = WCS("sky", "LINEAR", [1000,2000], [1, 1], [2, 2])
>>> x1, x0 = np.mgrid[1:3, 1:4]
>>> d = DataIMG("img", x0.flatten(), x1.flatten(), np.ones(x1.size), shape=x0.shape, sky=sky)
>>> print(d)
name = img
x0 = Int64[6]
x1 = Int64[6]
y = Float64[6]
shape = (2, 3)
staterror = None
syserror = None
sky = sky
crval = [1000.,2000.]
crpix = [1.,1.]
cdelt = [2.,2.]
eqpos = None
coord = logical
With this we can change to the “physical” coordinate system, which
represents the conversion sent to the sky argument, and so get the
independent axis in the converted system with the
set_coord method:
>>> d.get_axes()
(array([1., 2., 3.]), array([1., 2.]))
>>> d.set_coord("physical")
>>> d.get_axes()
(array([1000., 1002., 1004.]), array([2000., 2002.]))
>>> d.indep
(array([1000., 1002., 1004., 1000., 1002., 1004.]), array([2000., 2000., 2000., 2002., 2002., 2002.]))
It is possible to switch back to the original coordinate system (the
arguments sent in as x0 and x1 when creating the object):
>>> d.set_coord("logical")
>>> d.indep
(array([1, 2, 3, 1, 2, 3]), array([1, 1, 1, 2, 2, 2]))
In Sherpa 4.14.0 and earlier, this conversion was handled by taking
the current axes pair and applying the necessary WCS objects to create
the selected coordinate system (that is, the argument to the
set_coord call). This had the advantage of saving memory, as you
only needed to retain the current pair of independent axes, but at the
expense of losing fidelity when converting between the coordinate
systems. This has been changed so that the original independent axes
are now stored in the object, in the _orig_indep_axis attribute,
and this is now used whenever the coordinate system is changed. This
does increase the memory size of a DataIMG object, and makes it
harder to load in picked files created with an old Sherpa version (the
code will do its best to create the necessary information but it is
not guaranteed to work well in all cases).